Introduction
Materials and Methods
Plant materials
DNA extraction and PCR amplification
DNA sequencing and alignment
Phylogenetic analysis
Comparison of morphological characters
Results
Morphological characters
Phylogenetic analysis
Discussion
Morphological characters
Phylogenetic analysis
Naming of a new forma
Taxonomic treatment
Identification Key in O. monacantha complex
Introduction
At the 1578 Compendium of Materia Medica, Opuntia monacantha f. jejuensis was introduced as a green cactus in the Encyclopedia of Oriental Herbal Medicine and was studied until 1975. Since its introduction in Jeju, it has been used medicinally in the region (Kwon et al., 2017; Koh et al., 2018). Currently, O. humifusa is grown on a large scale in the Chungcheong and Jeolla regions (Kim et al., 2014). In particular, O. stricta is being cultivated on ~320 ㏊ of land (Yang et al., 2020). Cacti of the genus Opuntia, native to or grown in Korea, exhibit superior antibacterial, anti-glycosylation, antioxidant, anticancer, and cholesterol-lowering properties (Chung, 2000; Jung et al., 2012; Park et al., 2013; Choi, 2014; Jung et al., 2014). In addition to kimchi made with Opuntia powder, other foods, such as fermented milk, are also being made (Lee and Bae, 2009; Jung et al., 2016; Kim et al., 2016; Lee, 2017; Lee et al., 2018). Numerous studies have explored the potential of Opuntia as a cosmetic raw material (Lim and Hong, 2016; Kwon et al., 2017).
Cacti is a collective name for succulent plants belonging to the Cactaceae family of Caryophyllaes. In dry areas, during periods of heavy rainfall, water is stored in the plant body and used for plant growth during the dry season. Cactus has a reduced surface area to minimize water loss in arid regions, converting leaves to spines, and shortening stems and roots. Among the 3,000 species of succulent plants found worldwide, ~1,800 are cacti, which originated in South America and evolved in a dry environment (Anderson, 2001). Cactaceae are classified into four subfamilies by the International Cactaceae Systematics Group: Pereskoideae, Maihuenioideae, Opuntioideae, and Cactoideae (Nyffeler and Eggli, 2010). Opuntioideae is classified into seven genera: Brasiliopuntia, Tacinga, Consolea, Miqueliopuntia, Salmiopuntia, Opuntia and Tunilla (Wallace and Dickie, 2002; Griffith and Poter, 2009; Hernandez-Hernandez et al., 2011). In phylogenetic studies, it is classified into ten series: Brasiliopuntia, Tacinga, Elatae, Macbridei, Scheerianae, Humifusa, Macrocentra, Nopalea, Basilares and Microdasys (Table 1), (Majure et al., 2012b).
Table 1.
Classification of Opuntia s.s. based on phylogenetic studies (Majure et al., 2012b)
| Series | ||
|
Sister clade of Opuntia s.s. | Brasiliopuntia, Tacinga | |
| Opuntia s.s. | South American Clades | Elatae, Macbridei |
| North American Clades | Scheerianae, Humifusa, Macrocentra, Nopalea, Basilares, Microdasys |
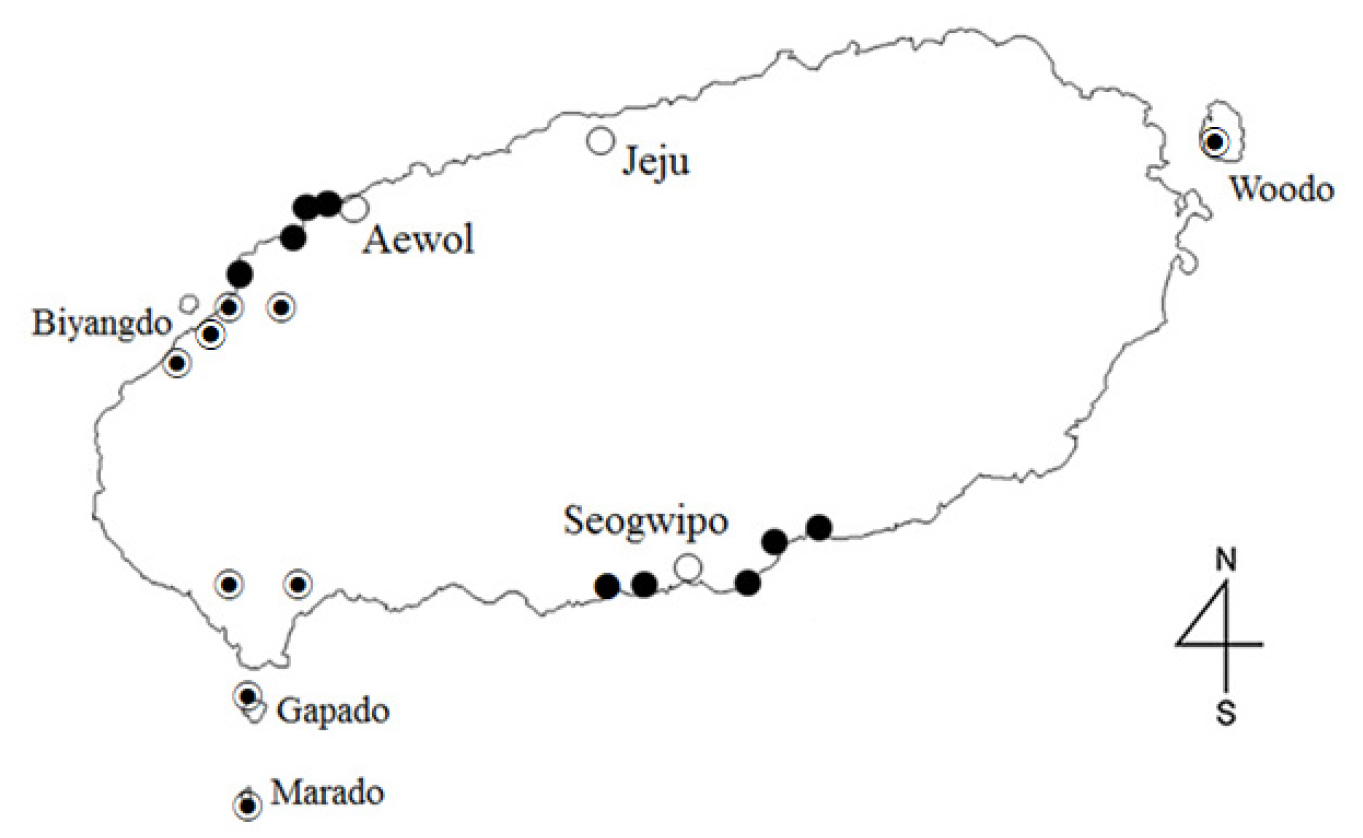
Opuntias.s. is a large genus of Cactaceae with ~200 species (Anderson, 2001). In Korea, O. humifusa and O. humifusa f. jeollaensis are native and are cultivated in the regions of Chungcheong, Jeolla, and Gyeongsang (Park et al., 2013; Kim et al., 2014). In the Jeju region, O. stricta Haw. also grows naturally and is cultivated (Yang et al., 2020; Yang and Oh, 2021), and O. monacantha f. jejuensis (this study) grows naturally (Fig. 1), (Yang and Oh, 2021).
O. monacantha f. jejuensis is the largest indigenous cactus species in Korea and is found on Jeju Island in the Aewol, Taepyeong, and Bomok regions (Fig. 1). In previous studies, External morphological studies have differentiated O. monacantha f. jejuensis from O. stricta by renaming the misidentified O. stricta, cultivated in the Hallim area of Jeju, and examining the phylogenetic relationship between them (Yang et al., 2020; Yang and Oh, 2021). The taxonomic analysis of O. monacantha sensu lato and its synonymous species is shown in Table 2. O. vulgaris Mill. had been accepted as a synonym of O. monacantha or O. humifusa but was later accepted as O. ficus-indica (L.) Mill. (Leuenberger, 1993). O. archavaletae Speg. was classified as a subspecies based on its different characteristics (flower, fruit) of reproductive organs (O.monacantha subsp. archavaletae (Speg.) Guiggi.) (Guiggi, 2017). A variety of O. monacantha was named O. monacantha var. gracilior Salm-Dyck (Table 2).
Table 2.
The taxonomic history of O. monacantha
Generally, phylogenetic analyses are conducted using the maximum likelihood (ML) and Bayesian inference (BI) methods which can be applied to a model of sequence evolution, and phylogenetic trees can easily be constructed using sequence data. Calculating the likelihood, ML maximizes the likelihood of the model for all data, whereas BI maximizes the posterior probability using likelihood. ML typically uses phylogenetic maximum likelihood (PhyML) or randomized accelerated maximum likelihood programs, whereas BI uses Bayesian inference of phylogeny (MrBayes) or Bayesian evolutionary analysis sampling tree (BEAST) programs to construct a phylogenetic tree. The obtained node support value for a phylogenetic tree written in ML is referred to as a bootstrap, and that in BI is called posterior probability. Majure et al. (2012a) used genetic ML and BI methods to conduct a phylogenetic analysis of the genus Opuntia. Kim et al. (2014) conducted genetic ML analysis and morphological classification by floral color for O. humifusa f. jeollaensis. Therefore, it was necessary to identify the position of O. monacantha f. jejuensis in the Opuntia via morphological comparisons and phylogenetic analysis using MI and BI methods.
Comparing the pulp color of O. monacantha f. jejuensis to that of O. monacantha, a distinct difference was observed in the characteristics of the two species (Koh et al., 2018; Yang and Oh, 2021). Owing to the lack of studies confirming the exact taxonomic position of O. monacantha f. jejuensis, which exhibits a distinct difference in pulp color and characteristics from O. monacantha, species identification studies are required before considering it as a raw material for pharmaceuticals, health supplements, and cosmetics. Therefore, to determine the exact phylogenetic position of O. monacantha f. jejuensis within the Elatae series, we compared the morphological characteristics of the O. monacantha complex and comfirmed its taxonomic status using phylogenetic analysis. This study contributes to validating a new forma of the taxon and its potential use as a health-promoting material.
Materials and Methods
Plant materials
The O. monacantha sensu lato used in this study was collected from the Taepyeong-ro habitat in Seogwipo (33°14'40.6" N 126°32'38.0" E). We transferred the collected materials were transferred to the cultivation site of the Baiknyuncho Museum in Hogeun-dong, Seogwipo, Jeju Island, for morphological studies. An immersion specimen was created and stored in Baiknyuncho Museum along with the associated resource management number. For DNA sequencing, one-year-old leaves were collected considering the occurrence of bacteria and contamination levels, and the obtained material was washed with distilled water, frozen in liquid nitrogen, and stored at -80℃ to extract the DNA.
DNA extraction and PCR amplification
Genomic DNA was isolated using the CTAB method (Doyle and Doyle, 1990). The primers and protocol were designed using the plastid intergenic spacers atpB-rbcL, trnL-F, and psbJ-petA, which are suitable for plant taxonomy phylogenetic classification studies, chloroplast gene matK, and nuclear ribosomal gene ITS (Table 3). The polymerase chain reaction (PCR) cycling parameters involved initial denaturation was performed at 94℃ for 5 min. The thermal cycle consisted of denaturation at 94℃ for 30 s, annealing at 55℃ for 30 s, and extension at 72℃ for 1 min, and was repeated 35 times. We performed the final extension at 72℃ for 10 min.
Table 3.
Genomic DNA regions and primer sequences used in this study
| Gene | sequence or reference | Length amplified | No. pars. infor. characters |
Model selected |
| atpB-rbcL | atpB Op : 5ˊ-GTAAACTATGTCGAAATTCTTTGC-3 ˊ rbcL. Op : 5ˊ-ACAACAAAACAACAAGGTCTACTC-3 ˊ | 861 | 20 | HKY |
| psbJ-petA | psbJ: (Shaw et al., 2007) petA. Op : 5ˊ-CAACATCAAGTTCGTAACAAG-3 ˊ | 1169 | 72 | K81uf+I |
| trnL-F | trnL: (Taberlet et al., 1991) trnF: (Taberlet et al., 1991) | 441 | 14 | K81uf |
| matK | matKx: 5ˊ-TAATTTACGATCAATTCATTC-3 ˊ matK5: 5ˊ-GTTCTAGCACCAGAAAGTCG-3 ˊ | 905 | 27 | F81+I+G |
| nrITS |
ITS4: (White et al., 1990) ITS5: (White et al., 1990) | 599 | 39 | TVM+G |
| cpDNA combined | - | 3407 | 115 | - |
| Nuclear combined | - | 568 | 57 | - |
| All combined | - | 3975 | 172 | - |
DNA sequencing and alignment
For sequencing of the five genes (atpB-rbcL, psbJ-petA, trnL-F, matK, and nrITS) of the investigated taxon, 25 cycles of sequencing reactions were performed using the Big Dye Terminator Cycle Sequencing Kit v.3.1 and the automated DNA sequencing system, Applied BioSystems DNA 3730XL Analyzer (Applied BioSystems, Foster City, CA, USA). DNA was purified using the Montage SEQ96 PCR clean-up kit (Millipore Corporation, Billerica, MA, USA). The nucleotide sequence was aligned using the ClustalX program (Thompson et al., 1997) and edited and rearranged using BioEdit ver. 7.2.6.2. program (Hall, 1999).
Phylogenetic analysis
After thoroughly confirming the DNA sequence of the target species, the sequences of the species were compared and analyzed from a molecular evolutionary perspective (Majure et al., 2012a,b). Three previously studied (Kim et al., 2014) cacti (O. humifusa, O. humifusa f. jeollaensis, and O. stricta) cultivated in Korea were sampled to obtain evolutionary data on O. monacantha f. jejuensis. Phylogenetic relationships were confirmed using the Tacinga series as outgroups using genetic information registered in the GenBank of the National Center for Biotechnology Information (Clegg, 1993; Soltis et al., 1998; Park et al., 2010), (Appendix 1). Comparative analysis was used to establish the exact position of O. monacantha f. jejuensis within the Elatae series and the form and DNA sequence of O. monacantha complex, including O. monacantha, O. monacantha subsp. arechavaletae, and O. monacantha f. jejuensis species (Appendix 2). BI and ML analyses were used to confirm the phylogenetic relationships between the species, and the programs MrBayes-3.2.5 (Ronquist et al., 2012) and PhyML 3.0 (Guindon et al., 2010) were used. The GTR+I+G (nst = 6, rates = invgamma) model, based on the main parameters affecting the substitution ratio in the DNA sequence, was used for BI analysis. Each of the five genes was analyzed using a random phylogenetic tree of 1×106 generations and 100 generations of Markov chains. In ML analysis, random samples were selected to duplicate the same number of characters as in the existing data set, repeated 10,000 times to form the same tree using PhyML 3.0 program, a phylogenetic tree, and bootstrap.
We constructed a molecular phylogenetic tree to illustrate phylogenetic relationships between species using TRE and PhyML files generated by BI and ML analysis, respectively. The molecular phylogenetic tree was confirmed using the FigTree v1.4.3 program (Rambaut, 2016), which was designed to display the summarized contents and annotated phylogenetic tree produced by BEAST.
Comparison of morphological characters
Six morphological characteristics, i.e., tepal color, stigma lobe color, fruit color, pulp color, and morphological differences between the seed, stem, and fruit, were selected through a comparative morphological study of the investigated taxon, i.e., O. monacantha complex, and evolutionary differentiation patterns within the O. monacantha complex were compared using a phylogenetic tree.
Results
The results of morphological and phylogenetic analyses of the Elatae series, which includes the O. monacantha complex, O. monacantha f. jejuensis, and Korean Opuntia spp., showed different flower colors, cladode shapes, fruit shapes, fruit colors, and pulp colors. O. monacantha f. jejuensis was assigned to the Elatae series in the phylogenetic analysis and was more closely related to O. monacantha subsp. arechavaletae than to O. monacantha at a molecular level.
Morphological characters
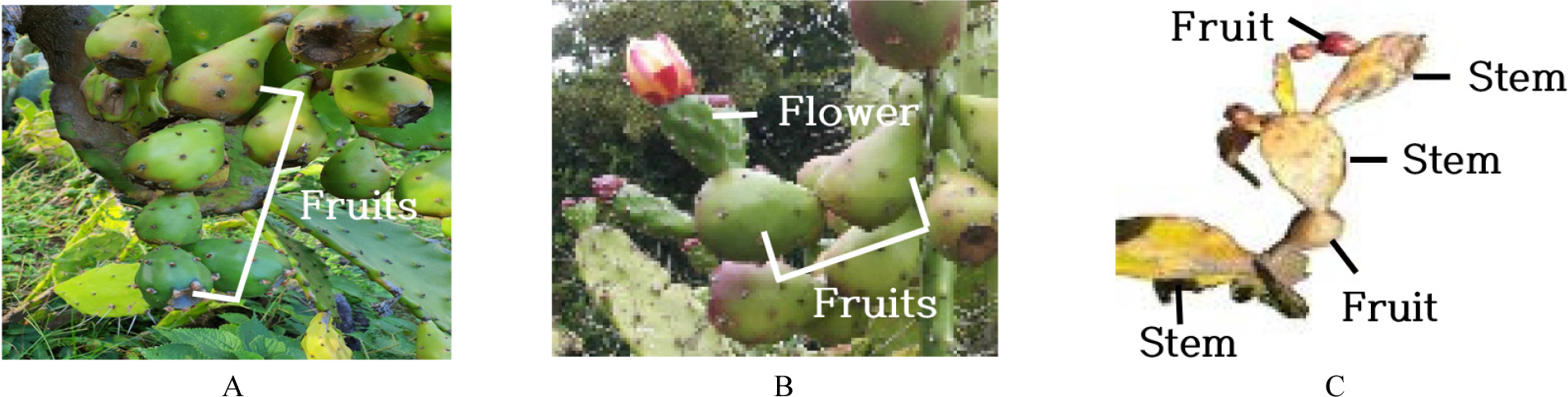
The investigated taxon had different morphological characteristics than other Opuntia spp. in Korea (Table 4). The cladodes of mature specimens of O. stricta, O. humifusa, and O. humifusa f. jeollaensis are ovate, but those of O. monacantha f. jejuensis were oblong. Furthermore, plant size, flowering period, fruit shape, and pulp color were significantly different. O. monacantha f. jejuensis is a tree-like plant that grows to a height of > 2 m, while other species are shrub-like plants that grow to a height of GTTCTAGCACCAGAAAGTCG 1 m. O. monacantha f. jejuensis fruits are plum-shaped, whereas those of other species are elongated pear-shaped. The fruit of the investigated taxon is purple-green, whereas those of the others are reddish-purple (Fig. 2). Finally, the pulp of the investigated taxon was yellow-green, whereas that of the other species was purple.
Table 4.
Morphological differences of O. monacantha f. jejuensis and Opuntia spp. in Korea

Fig. 2.
Morpholological characteristics of Cladode, flowers, fruit, and pulp of O. monacantha f. jejuensis, illustrating their morphological characteristics. The scale bar (white) across the bottom of each photograph is 2 ㎝. O. monacantha f. jejuensis (A). Germinated O. monacantha f. jejuensis (B). Oblong shape of a cladode (C). Flower with purple stripes on the outer surface of yellow petals (D,E). Plum-shaped fruit (F-H). Spine (4 ㎝ long) (I). Stigma lobes (×10) (J).
O. monacantha f. jejuensis was observed to be morphologically similar to O. monacantha. However, O. monacantha had yellow tepals with abaxially red midveins, i.e., purple or red stripes on the front and back of tepals (Fig. 3), (Majure and Puente, 2014), whereas these stripes appear only on the back of O. monacantha f. jejuensis tepals. O. monacantha and O. monacantha subsp. arechavaletae had yellow or orange-yellow tepals, whereas O. monacantha f. jejuensis had bright yellow tepals (Fig. 3).

Fig. 3.
Flower and fruits of O. monacantha complex.
Flower of O. monacantha (A-1): https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:323260-2/images
Fruits of O. monacantha (A-2): https://www.gbif.org/occurrence/3859139935
Flower of O. monacantha subsp. arechavaletae (B-1) https://www.gbif.org/occurrence/3759979394
Fruits of O. monacantha subsp. arechavaletae (B-2) https://www.gbif.org/occurrence/3759979394
Stigma lobe color, fruit color, seeds, pulp color, and peculiar phenomenon were the traits used for morphological classification. O. monacantha has creamish stigma lobes, O. monacantha subsp. arechavaletae has greenish stigma lobes (Guiggi, 2017; Maria et al., 2017), and O. monacantha f. jejuensis has creamish stigma lobes (Table 5, Fig. 3). The mature fruit of O. monacantha and O. monacantha f. jejuensis is purple-green, whereas those of O. monacantha subsp. arechavaletae is reddish-purple. The filaments of the stamens of O. monacantha were greenish in color, according to previous studies (Pardo and Alonso, 2017), but in this study they turned out to be yellow. Those of O. monacantha f. jejuensis were yellow, too. Sprouts of O. monacantha f. jejuensis were germinated (Fig. 2). The fruiting pattern on stems in the investigated taxon can be classified into two distinct types. One is that a stem grows from a fruit, with another fruit growing from this stem and the other is that three fruits grow sequentially at the end of a single stem, which is the most characteristic phenomenon that occurs only in O. monacantha f. jejuensis (Fig. 4).
Table 5.
Morphological differences in O. monacantha complex
| Taxa | O. monacantha | O. monacantha subsp. arechavaletae | O. monacantha f. jejuensis |
|
Flower color |
Yellow or orange yellow tepal, Red stripes on the inside and outside of tepals |
Yellow or orange yellow tepal, Red stripes on the outside of tepals |
Yellow tepal, Red stripes on the outside of tepals |
|
Stigma lobes color |
Pale yellow to pale creamish (Haworth, 1819) |
Greenish (Guiggi, 2017) | Pale yellow to pale creamish |
| Fruit color |
The fruit maturing and reddish purple, greenish near base or green with red-purple shades (Haworth, 1819) |
The fruit maturing and reddish-purple, greenish near base (club head part) |
The fruit maturing and reddish purple, greenish near base or green with red-purple shades |
| Fruit shape |
Narrowly turbinate to obovoid fruits, with well-developed loculus (Maria et al.,2017) Plum shape |
Longer, elongate, with an apical and smaller loculus (Guiggi, 2017) Club shape |
Narrowly turbinate to obovoid fruits, with well-developed loculus Plum shape |
| Seeds |
3.5×4 mm (Haworth, 1819) | - |
Seeds are light tan, Irregularly elliptic, ca. 2 ea 5×6 mm More than 50 ea 3×4 mm |
| Pulp color |
Greenish (Maria et al., 2017) |
Greenish (Maria et al., 2017) |
Inside: Greenish Outside: Green-yellow |
|
Peculiar phenomenon | - | - |
Fruit+Stem+Fruit, Fruit+Fruit+Fruit |
Phylogenetic analysis
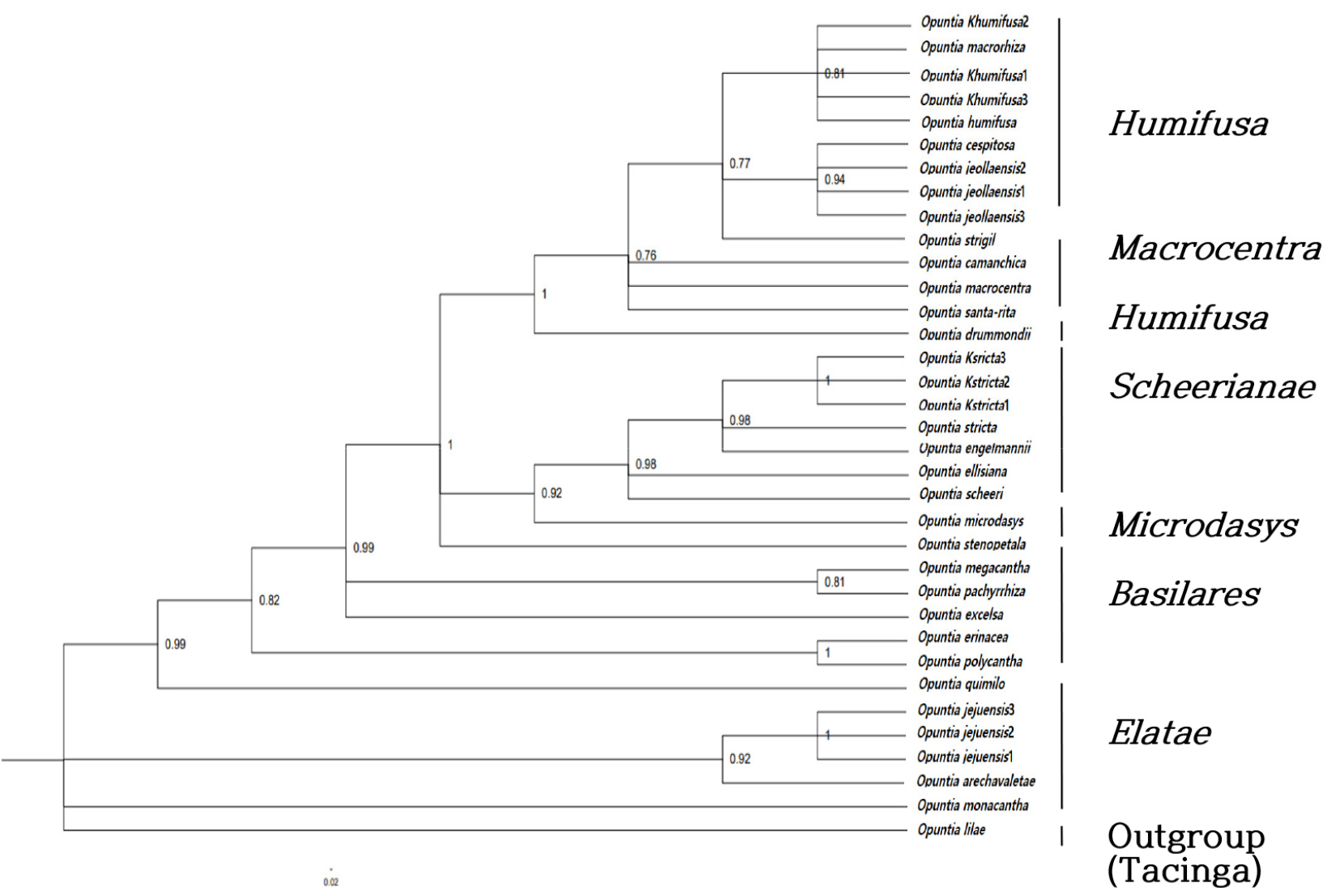
Bayesian trees based on DNA sequence showed posterior probabilities summarized from a set of post-burn-in trees generated using the GTR+I+G model (Figs. 5 and 6). Opuntia stricta was included in Scheerianae series and O. humifusa and O. humifusa f. jeollaensis were included in Humifusa series (Fig. 5). These three species formed a part of North American clades. However, unlike other Opuntia spp. in Korea, O. monacantha f. jejuensis was included in Elatae series of South American clades (Fig. 6) as it shares numerous morphological characteristics with O. monacantha. The phylogenetic distinctions between O. monacantha f. jejuensis, O. monacantha, and O. monacantha subsp. arechavaletae (Fig. 6) were not distinguished. We conducted ML and BI analyses of the O. monacantha complex and other species in South American clades to identify the detailed molecular evolutionary aspects of O. monacantha f. jejuensis. The results of the analyses were highly relevant, because O. monacantha f. jejuensis branched with O. monacantha subsp. arechavaletae within O. monacantha complex.

Fig. 6.
Bayesian and ML trees of South American clades including O. monacantha complex taxa (using atpB-rbcL, trnL-F, psbJ-petA, matK and nrITS sequences). BPP (right) and Bootstrap values (left) are shown between branches. South American clades used the names of the series published by Britton and Rose (1920), Engelmann (1856), and Majure et al. (2012a) as a result of their research. The BPP values are > 0.6 and The Bootstrap values are ≥ 60%.
Discussion
Morphological characters
When compared to O. monacantha (Wagner et al., 1999; Taylor and Zappi, 2004; Navie and Adkins, 2008) and O. monacantha subsp. arechavaletae (Speg, 1905; Guiggi, 2017), O. monacantha f. jejuensis (Yang and Oh, 2021) showed significant differences in flower color, stigma lobe color, fruit color, seed color, pulp color, and peculiar phenomenon (Fig. 4, Table 4). Based on the findings of this study, O. monacantha f. jejuensis has a pulp color that is distinct from that of O. monacantha. Yang and Oh (2021) reported that O. monacantha f. jejuensis, a species native to Jeju Island, was morphologically different from O. monacantha in fruit and stem growth characteristics. As the mature fruits of plants in Opuntia have the same color as pulp, fruit color indicates pulp color (Omweri et al., 2016). However, the pulp of O. monacantha f. jejuensis is greenish-yellow, which differs from the color of its fruit (Table 5). The mature fruit of O. monacantha and O.monacantha f. jejuensis are purple-green, whereas that of and O. monacantha subsp. arechavaletae are reddish-purple. Moreover, O. monacantha f. jejuensis exhibited an unusual flowering phenomenon on the fruit. In other plants, it is common for the fruit to grow on the stem, but O. monacantha f. jejuensis also showed stem growth from in the fruit. Therefore, fruit areole has the same function as stem areole. This study showed that O. monacantha f. jejuensis is morphologically different from O. monacantha and thus can be classified as a new forma.
Phylogenetic analysis
Following Kim et al. (2014), wherein plastid intergenic spacer trnL-F, plastid gene matK, and nrITS, were used, this study included the three taxa, O. stricta, O. humifusa, and O. humifusa f. jeollaensis, in North American clades. O. stricta was included in Scheerianae series and O. humifusa and O. humifusa f. jeollaensis were included in Humifusa series (Majure et al., 2012b). O. monacantha f. jejuensis was included in Elatae series of South American clades, which is consistent with the findings of previous studies (Yang and Oh, 2021) that it is evolutionarily closer to O. monacantha subsp. arechavaletae than to O. monacantha. Furthermore, according to the research findings of Majure et al. (2012a,b), Kim et al. (2014), and Yang and Oh (2021), alloploid species are a radial evolution type in North American clades. When comparing the O. monacantha complex with other species in Elatae series using phylogenetic trees based on plastid and nrITS sequences, the results were consistent with previous findings that O. monacantha f. jejuensis was evolutionarily closer to O. monacantha subsp. arechavaletae than O. monacantha.
Naming of a new forma
Recently, O. monacantha subsp. arechavaletae has been classified as a subspecies of O. monacantha (Guiggi, 2017). The stigma lobes are yellow in O. monacantha but green in O. monacantha subsp. arechavaletae. O. monacantha f. jejuensis differed morphologically from O. monacantha in several areas (Table 5). The fruit shape, size, stem, and spines of the investigated taxon were morphologically similar to those of O. monacantha, making it difficult to regard it as a new species. However, in light of the morphological (flower color, pulp color, and peculiar phenomenon) and phylogenetic (phylogenetic trees) analysis of O. monacantha f. jejuensis, it was deemed reasonable to classify it as a new forma. The species was named O. monacantha (Willd.) Haw. f. jejuensis J. K. Kim ex Y. S. Yang, based on the habitat characteristics, i.e., native or widely cultivated in Jeju, and the first discoverer and scientific name designator was Jekuk Kim, who is a strong advocate for the taxon's protection, cultivation, and breeding.
Taxonomic treatment
Opuntia monacantha (Willd.) Haw.f. jejuensis J. K. Kim. ex Y. S. Yang,for. nov. (Fig. 7)
Korean name: Je-ju-baik-nyun-cho 제주백년초
Origin: Eastern coastal South America (Argentina, Brazil, Paraguay, and Uruguay)
A type of succulent tree (thorny shrub); Erect cylinder trunk shrub, trunk diameter 25-30 ㎝; Cladodes shiny green, 2-5 m height, oblong (elongated), 20-35 ㎝ long, 8-12 ㎝ wide, areoles arranged > 2 ㎝ wide; Leaves subulate; Petals always yellow with purple median stripes on the outer perianth segment, 16-25 mm long and 11-16 mm ; Bright yellow inner perianth parts, 22-40 mm long and 16-42 mm wide, with yellow to white staminal filaments (180-200 ea), the style yellow, 10-20 mm long, divided into 5-6 bright yellow stigma lobes and 8-10 mm long; Flowering May-January; Reddish flower buds; Spines, 4-5 ㎝ long, white-grey or yellowish to reddish-brown; Brownish glochids numerous; Seeds, irregularly elliptic, usually 6×5 mm 2 ea and 4×3 mm More than 40 ea, High germination rate (> 90%); Fleshy fruit, purple- green, yellow-green pulp color, plum-shaped, 5-7.5 ㎝ long and 4-5 ㎝ in diameter; Fruiting June to next year April.
Diagnostic characters: Red stripes on the outside of petals, yellow stigma lobes, purple-green fruit, plum-shaped fruit, green-yellow pulp, peculiar phenomenon (securely generated fruit, flower growing from fruit, stem growing from fruit).
Holotype: Taepyeong-ro 200, Seogwipo, Jeju Province, Korea. (33°14'40.6" N 126°32'38.0" E) Oct. 17, 2020. (YANG-202201) Herbarium of Jeju Baiknyuncho Museum. Donation to Herbarium of Jeju Baiknyuncho Museum (JBM).
Isotype: Taepyeong-ro 200, Seogwipo, Jeju Province, Korea. (33°14'40.6" N 126°32'38.0" E) Oct. 17, 2020. (YANG-202202, YANG-202203, YANG-202204) Herbarium of Jeju Baiknyuncho Museum (JBM).
Distribution: Jeju Island, Korea
Etymology: The specific epithet was derived from Jeju Island, where this new taxon is located.
Habitats:O. monacantha f. jejuensis grows on the coast of Seogwipo-si, Jeju-si, Jeju Province, Korea
Identification Key in O. monacantha complex
1a Yellow-orange or orange flowers Elatae series
1b Yellow or Yellow-orange flowers 2(O. monacantha complex)
2a Greenish stigma lobes O. monacantha subsp. arechavaletae
2b Creamish stigma lobes 3(O. monacantha subsp. monacantha)
3a Red stripes inside and outside tepals O. monacantha f. monacantha
3b Red stripes on the outside of tepals O. monacantha f. jejuensis